Research Skills
High-dimensional & multi-modal data analysis
- In my Ph.D., I analyzed high-dimensional single-cell datasets using sparse
cell x genematrices with thousands of individual features and up to tens of millions of unique cells:
Gene_1 Gene_2 ⋯ Gene_m Cell_1 ⋯ ⋯ ⋯ ⋯ Cell_2 ⋯ ⋯ ⋯ ⋯ ⋯ ⋯ ⋯ ⋯ ⋯ Cell_n ⋯ ⋯ ⋯ ⋯
-
These datasets contained multiple
cell x featurematrices with the multi-modal profiles for each cell -
To analyze these datasets, I used unsupervised machine-learning methods such as t-SNE and Louvain clustering:
Reagor & Hudspeth, 2024, bioRxiv
Deep learning for causal time-series analysis
-
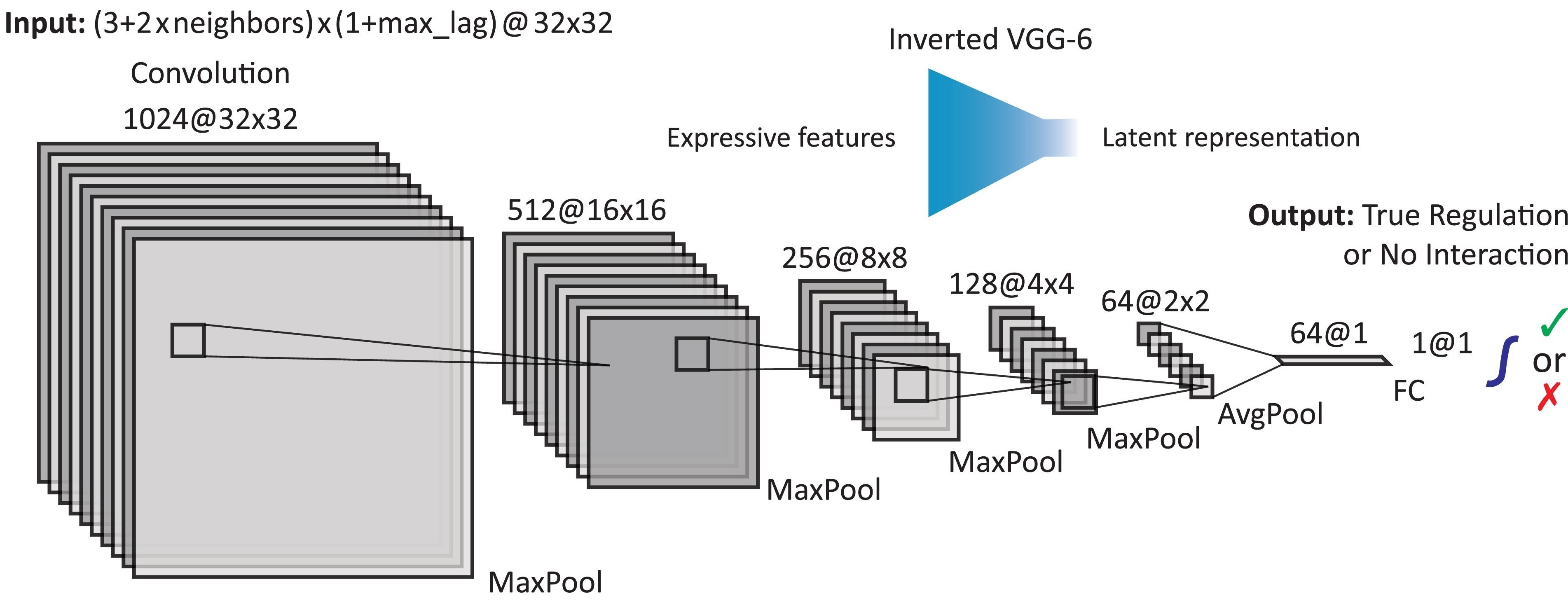
I developed the deep-learning method DELAY to reconstruct causal gene-regulatory networks from single-cell gene-expression datasets
-
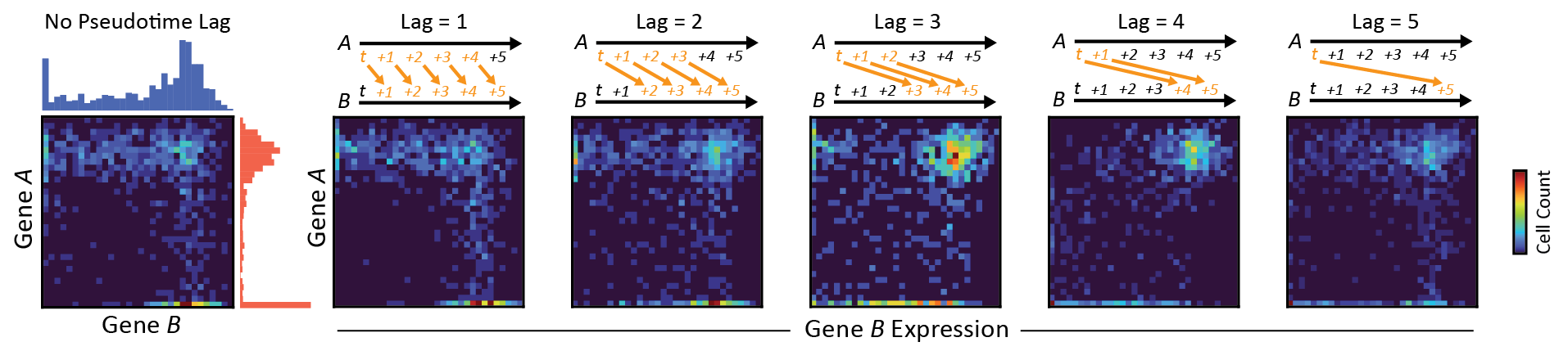
Using concepts from Granger Causality, I designed DELAY to encode noisy gene-expression data as images for deep learning
Reagor, Velez-Angel & Hudspeth, 2023, PNAS Nexus
- DELAY uses a convolutional neural network to classify images as either interacting or non-interacting gene pairs
Analysis of large-scale networks
-
To reconstruct large-scale networks, I inferred causal interactions between small clusters of genes
-
I used graph theory to identify the nodes (genes) that control the network’s temporal dynamics
To validate this reconstructed network, I performed experiments on regenerating zebrafish
- Beyond DELAY, I also developed custom scripts to quantify and analyze neuronal networks